Proteomics Journal
Objective:
The objective of this document is to journal the process and findings related to proteomics data analysis.
Tools & Technologies used:
1. Detection of ORF (NCBI BLAST): NCBI BLAST was utilized to detect Open Reading Frames (ORFs) within the proteomics data.
2. Phylogenetic analysis: Phylogenetic analysis software, such as NCBI Phylogeny, was used to analyze the evolutionary relationships among the identified proteins.
3. Prediction of the secondary structure of protein: Tools like J-Pred 4 were employed to predict the secondary structure elements (e.g., alpha helices, beta sheets) of the proteins.
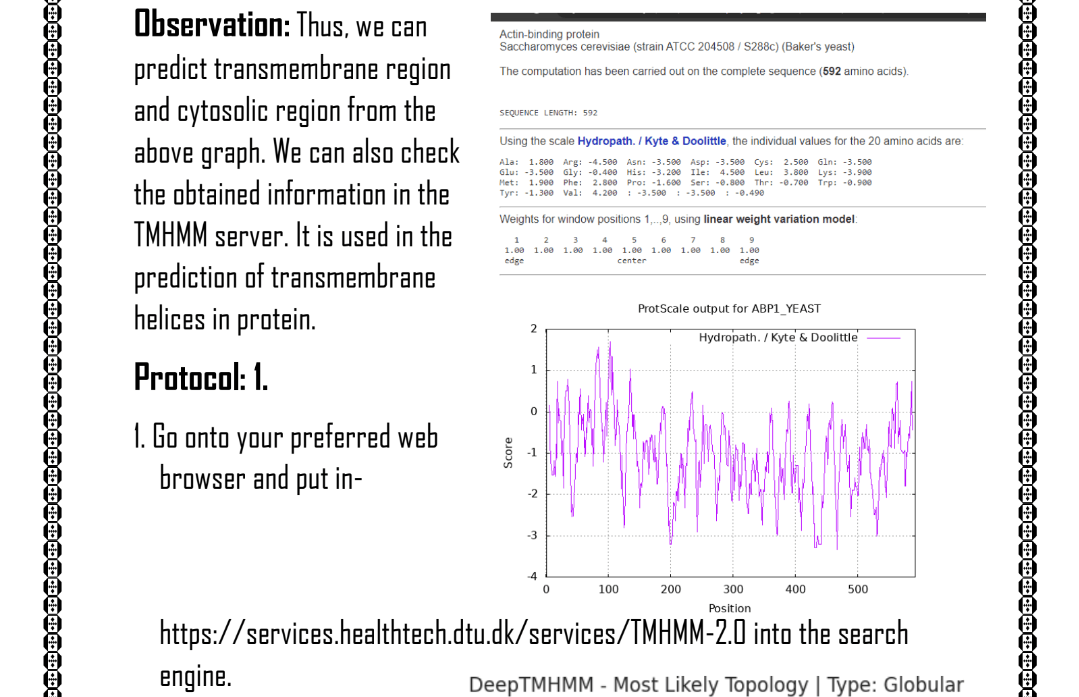
4. Hydropathy plot: Hydropathy plot tools, such as Kyte-Doolittle or GRAVY, were used to analyze the hydrophobicity patterns of the identified proteins.
5. SDS-PAGE: Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) was performed to separate and visualize the proteins based on their molecular weight.
Target Keywords:
The targeted keywords for this study encompassed proteomics, data analysis, ORF detection, NCBI BLAST, phylogenetic analysis, secondary structure prediction, hydropathy plot, and SDS-PAGE.
Challenges and learning:
Throughout the project, several challenges were encountered, such as optimizing the ORF detection parameters, interpreting phylogenetic tree topologies, refining secondary structure predictions, analyzing complex hydropathy patterns, and optimizing SDS-PAGE conditions. These challenges provided valuable learning opportunities, enhancing the understanding of proteomics data analysis and the interpretation of different analysis outputs.
Result/Outcome:
The project yielded significant outcomes, including the identification and annotation of ORFs, elucidation of evolutionary relationships among proteins, prediction of secondary structure elements, characterization of protein hydrophobicity patterns, and visualization of protein separation by SDS-PAGE. These results contribute to a better understanding of protein structure, function, and evolutionary relationships in the context of proteomics research.
28 Mar 2023